Compound-specific stable isotope analysis (CSIA) has seen an explosion of publications in recent years across diverse fields of ecology, oceanography, and biogeochemistry. The power of the CSIA approach lies in the differential fractionation of individual compounds (e.g., amino acids) during trophic transfer. In this way we can disentangle the relative influences of trophic variability and base of the food web variability on consumer stable isotope values. Furthermore, in contrast to many extant biomarkers, amino acids are major biochemical constituents of all organic matter, accounting for approximately half of the total organic carbon and most of the organic nitrogen in organisms. Therefore, amino acids are major conduits of carbon and nitrogen flow through food webs. As such, CSIA has opened the doors for more detailed examinations of a wide range of interconnected topics, including resource utilization and trophic dynamics, animal migration, and biogeochemical cycling.
One major component of my research is focused on understanding the patterns of stable carbon and nitrogen isotope trophic fractionation. I am particularly interested in exploring the underlying biochemical and physiological mechanisms of this fractionation, both as a tool to understand how consumers utilize and modify resources, and as a tool to advance the field of compound-specific stable isotope analysis. Much of this work is done through controlled feeding experiments where we examine isotopic offsets between diet and consumer of a wide range of organisms. The ultimate goal of this aspect of my research is to provide the analytical foundation for more applied research on how consumer-resource relationships influence the structure, function, and resilience of complex ecosystems.
Carbon Isotopes
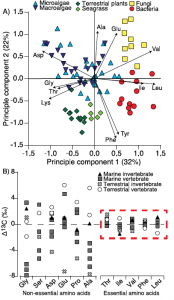
A) Amino acid fingerprinting, via PCA, showing separation of phylogenetic groups based on normalized essential amino acid d13C values (Larsen, et al. 2013) and B) amino acid d13C
change with trophic transfer (D13C) showing high variability across non-essential amino acids but little to no fractionation for essential amino acids across a range of marine and terrestrial invertebrate and vertebrate consumers (modified from McMahon et al. 2013).
Individual amino acids are typically divided into two main categories with respect to their carbon isotope fractionation during trophic transfer, essential and non-essential amino acids. While plants, algae, and bacteria can synthesize essential amino acids de novo from a bulk carbon pool, animals have lost the enzymatic capability to synthesize these amino acids at a rate sufficient for normal growth. As such, animals must acquire their essential amino acids directly from their diet with little to no trophic fractionation. Furthermore, the d13C composition of organisms varies widely (~20‰) among individual amino acids, both in absolute value and relative d13C pattern, reflecting the sum of the isotopic fractionations associated with the individual biosynthetic pathways and associated branch points for distinct amino acids. There is a tremendous amount of evolutionary diversity within the central biosynthetic amino acid pathways, which results in diagnostic amino acid d13C patterns linked to carbon acquisition and metabolism. The d13C patterns of essential amino acids are particularly diagnostic of phylogenetic source identity, in other words, the type of primary producer that made those amino acids. This is because essentials amino acids have very long and complex biosynthetic pathways (five or more biosynthetic steps) compared to many other biomolecules, providing far greater potential for lineage-specific isotope effects. As a result, essential amino acid isotope patterns provide an isotopic fingerprint of the phylogentic identity of sources at the base of food webs that are passed on through food webs virtually unmodified. Much of my work on carbon isotope fractionation in amino acids is focused on understanding what drives the separation in amino acid d13C values among primary producers and using that knowledge to develop more refined and robust amino acid fingerprints of dietary reconstructions. This work is currently being funded by a Packard Endowment Research Grant.
In contrast to the generally well characterized essential amino acids, the underlying drivers of variability in non-essential amino acid fractionation are still largely unclear. Non-essential amino acids can either be de novo biosynthesized from a bulk carbon pool or directly routed from dietary protein, with typically highly variable D13CC-D values across taxa and diet types. Some recent work from my lab as well as several close colleagues suggest that non-essential amino acid fractionation is likely related to diet composition and quality. I believe that the stable carbon isotope values of non-essential amino acids may hold fascinating clues to how consumers utilize complex heterogeneous diets. My hope is that in the future we may be able to use these signals to quantify diet quality and macromolecular metabolism in wild animals.
Nitrogen Isotopes
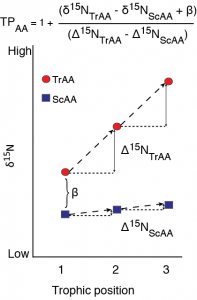
A schematic of fractionation in individual amino acid nitrogen isotope value with trophic transfer. Trophic amino acids (TrAA, e.g., glutamic acid) show large fracitonation with each trophic transfer while source amino acids (SrAA, e.g., phenylalanine) show little to no fractionation with trophic transfer. Together, these amino acids can provide an estimate of consumer trophic position that is internally normalized to the baseline nitrogen isotope value of the food web.
With respect to nitrogen, individual amino acids are commonly divided into trophic and source categories based on their relative fractionation with trophic transfer. Trophic amino acids, most commonly represented by glutamic acid (Glu), undergo significant isotopic fractionation during transamination/deamination. These amino acids provides greater sensitivity for defining trophic position than bulk stable isotope analysis. Conversely, the canonical source AA, phenylalanine (Phe), shows minimal trophic fractionation between diet and consumer because its dominant metabolic processing does not form or break C-N bonds. Thus Phe d15N provides a proxy for the sources and cycling on nitrogen at the base of food webs (d15Nbaseline). Together, the d15N value of Glu and Phe can be used to estimate consumer trophic position while accounting for differences in d15Nbaseline without needing to independently characterize and analyze the baseline structure of a food web. The accuracy of this approach fundamentally depends on the accuracy of the trophic discrimination factor used to calculate trophic position. Accumulating evidence, however, suggests that TDFGlu-Phe values may in fact vary widely across taxa, likely related to diet quality and mode of nitrogen excretion. I currently have several related projects going that look to 1) quantify the variability in amino acid nitrogen isotope fractionation between diet and consumer across a wide range of species and diet types, 2) explore the underlying biochemical and physiological mechanisms generating the variability in isotope fractionation, and 3) develop new trophic dynamics models to accurately incorporate this variability in trophic position estimates. Together this work will pave the way for more accurate and robust studies of trophic dynamics in natural systems.
Publications:
McMahon KW, McCarthy MD (In Press) Embracing variability in amino acid d15N fractionation: Mechanisms, implications, and applications for trophic ecology. Ecosphere
McMahon KW, Elsdon T, Thorrold SR, McCarthy M (2015). Trophic discrimination of nitrogen stable isotopes in amino acids varies with diet quality in a marine fish. Limnology and Oceanography 60:1076-1087